
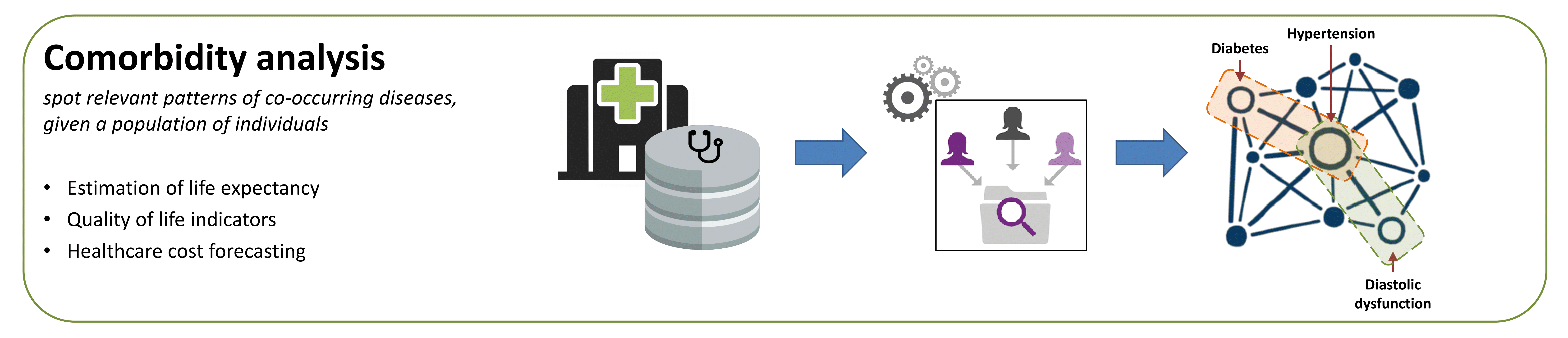
Overview of Comorbidity4j
Comorbidity4j is a Java tool that efficiently carries out comorbidity analysis over patient-dagnosis datasets.
Any kind of user interaction in Comorbidity4j is carried out by means of a Web interface: this Web interface drives users in providing and validating input data, defining the parameter of the comorbidity analysis and accessing or downloading its results.
Comorbidity4j Web interface can be accessed:
- on-line: at http://comorbidity.eu/comorbidity4web/ by means of the Comorbidity4web service;
- locally on the user workstation: by downloading the Comorbidity4j Java library and locally executing the Comorbidity4j server. Instructions for local execution of comorbidity analyses can be accessed at Executing Comorbidity4j on your pc. Comorbidity analyses executed by Comorbidity4j expects the same input data and produce the same set of interactive Web visualizations generated by Comorbidity4web, but data are processed and results are stored locally to the user workstation. In this way it is possible to enforce a greater data privacy, protect sensitive data or exploit more powerful workstations.
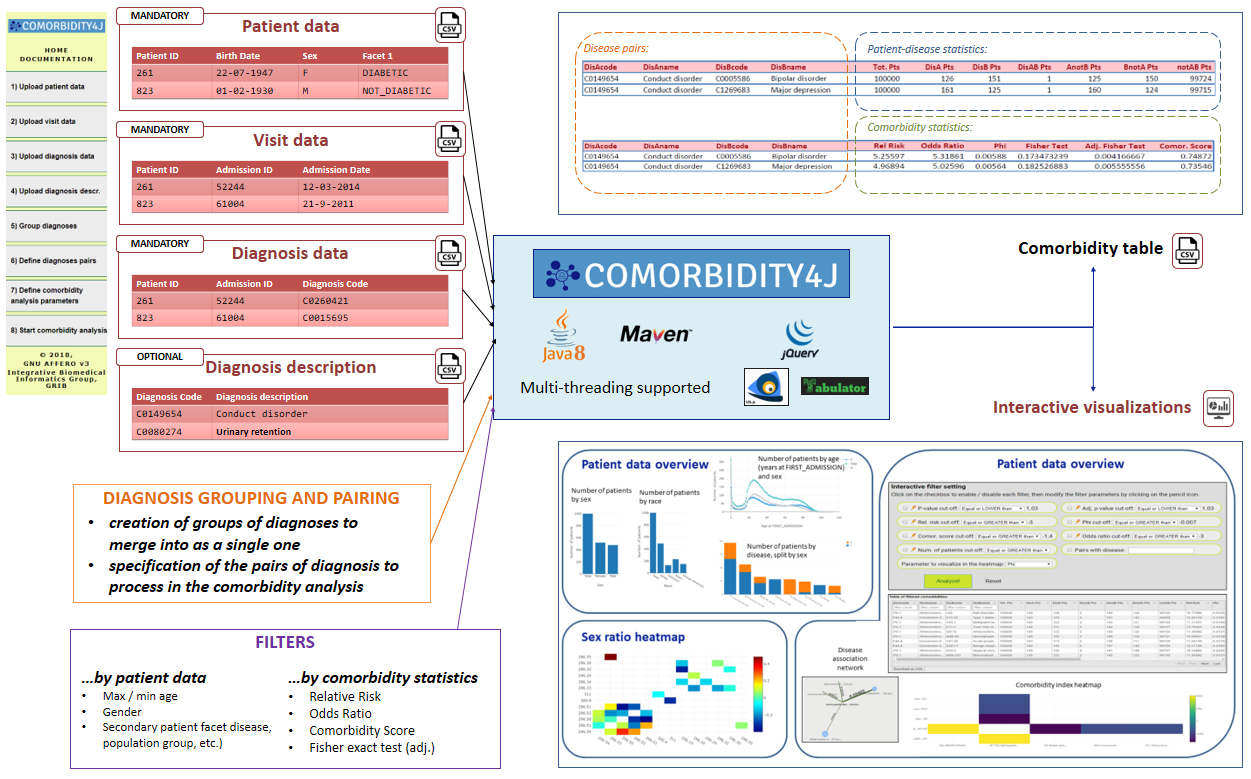
Input data are provided by a set of spreadsheets useful to describe patients and their diagnoses (referred to as Patient input files). These files are interactively uploaded and validated by means of the Web interface of COmorbidity4j. To get more details, go to Prepare your comorbidity analysis data (section below).
Once you have uploaded your input data, validated them and set the comorbidity analysis parameters, you can execute a comorbidity analysis (section below). Comorbidity analyses over small-to-medium sized patient-disease datasets can be executed online by means of the Comorbidity4web service, otherwise you can download Comorbidity4j and execute it on your PC (see Executing Comorbidity4j on your pc).
The results of an analysis of comorbidity includes:
- a Comorbidity table that provides for each pair of diseases a set of Comorbidity scores
- a set of Web-based interactive visualizations useful to explore several facets of the input patient-disease dataset as well as to apply further filters on the results of comorbidity analyses and visualize these results by means of interactive heatmaps and network graphs. From this link you can access an example of the interactive Web visualizations generated by Comorbidity4j.
The following image visually shows the main capabilities of Comorbidity4j:
Prepare your comorbidity analysis data
Comorbidity4j takes as input a set of files, referred to as Patient input files and describing the demographic information (birth-date, gender and, optionally secondary patient features like education level, ethnicity, etc.) and the history of diseases of a set of patients. See: Patient input file format for detailed information on the format of each file.
It is possible to download an example dataset (generated by Synthea) to inspect the input tabular files that Comorbidity4web expects as input to execute a comorbidity analysis and try the tool.
In general, the set of Patient input files includes the following four spreadsheets:
- Patient data file: (MANDATORY, download example) list of patients, one per row. Each patient is described by a unique identifier and a set of demographic information (birth-date, gender and, optionally secondary patient features like education level, ethnicity, etc.)
- Visits data file: (MANDATORY, download example) list of visits (admissions) of a patient. Each visit of a patient is described by a unique identifier, the unique identifier of the patient who was attended and the date of the visit
- Diagnosis data file: (MANDATORY, download example) list of diagnoses (diseases) that were discovered for a patient in the context of a specific visit. Each diagnosis includes the unique identifier of the patient, the unique identifier of the visit and the unique identifier of the diagnosis
- Diagnosis description file: (OPTIONAL, download example) list of extended free text description of unique identifiers of diagnoses. Each row associates the unique identifier of a diagnosis to an extended free text description of the same disease. If this file is not provided, each disease will be referenced in the results of the comorbidity analysis by its unique identifier (that could be a code (ICD-9, SNOMED ID) from which it is difficult to derive the semantics of the disease
Comorbidity4j can read Patient input files in a format compliant with the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) as described into details at Processing OMOP Common Data Model datasets.
Execute a comorbidity analysis
Comorbidity4j can be executed in any PC where Java 1.8 or newer is present (or online at http://comorbidity.eu/comorbidity4web/). Once connected by means of a Web browser to Comorbidity4j (to a locally running server to to http://comorbidity.eu/comorbidity4web/), input data (see Prepare your comorbidity analysis data) can be interactively uploaded and validated, analysis parameters specified and comorbidity analysis executed. See Executing Comorbidity4j on your pc to get detailed instruction on how to download and execute Comorbidity4j on your PC.